https://meetinglibrary.asco.org/record/198553/abstract
Authors:
Toshiki Masuishi, Hiroya Taniguchi, Daisuke Kotani, Hideaki Bando, Taroh Satoh, Taito Esaki, Yoshito Komatsu, Yu Sunakawa, Tomohiro Nishina, Eiji Shinozaki, Naohiro Nishida, Masato Komoda, Satoshi Yuki, Naoki Izawa, Gaurav Sharma, Stan Skrzypczak, Eric Schultz, Carl Kingsford, Akihiro Sato, Takayuki Yoshino
Research Funding:
Aichi Cancer Center Hospital
Background:
Patients (pts) with BRAF V600E mutant (mt) metastatic colorectal cancer (mCRC) still have a poor prognosis even when treated with encorafenib plus cetuximab. We reported that eribulin that inhibits G2-M cell cycle had limited activity for these pts in the phase II BRAVERY study (Masuishi T, et al. WCGC 2020). Barras D, et al. (2017) reported that BRAF V600E mt mCRCs were classified into BM1 and BM2 characterized by KRAS/AKT pathway activation and deregulation of the cell cycle with G2-M phase activation, respectively, based on gene expression.
Methods:
Whole transcriptome RNA-seq of FFPE pre-treatment tumor samples was performed using NovaSeq 6000 in the BRAVERY study. Molecular features were extracted using the Txome.ai platform (Ocean Genomics Inc., PA, USA) which included transcript- and gene-level expression quantification, expression-based clustering, and structural variant calling. Efficacy of eribulin was classified as “good” if pts had a tumor reduction and/or progression-free survival (PFS) of more than 6 months, and “poor” otherwise. The differential gene expression analysis was performed between pts with “good” and “poor” using Txome.ai. In addition, BM and consensus molecular subtype (CMS) classification were performed using the model developed by Barras D, et al. (2017) and Guinney J, et al. (2015), respectively.
Results:
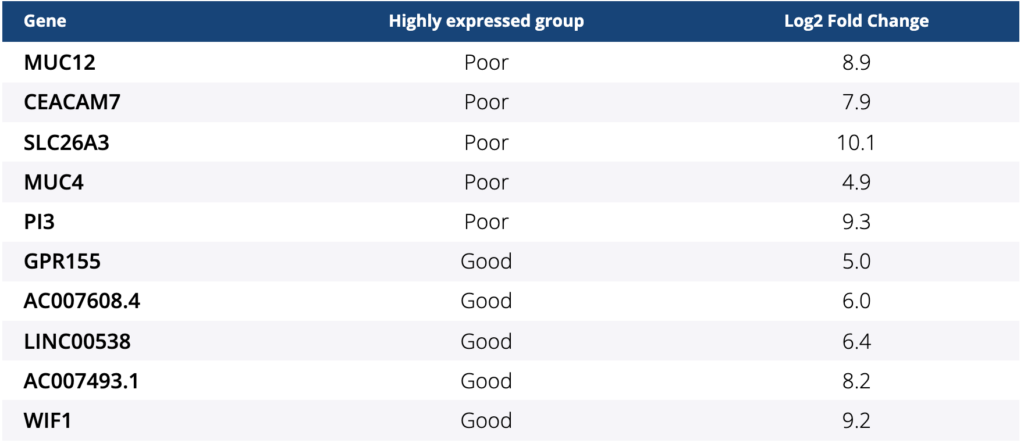
Among 27 pts, 26 tumor samples were available to perform RNA-seq and analyze gene expression despite low mapping rates. Patient characteristics were as follows: median age of 58.5 (range, 33–71) years; ECOG PS of 0/1 (16/10); primary tumor location of right/left (11/15); and all 26 pts had MSS/pMMR. Four and 22 pts were classified into “good” and “poor” groups, respectively. Among 52 differentially expressed genes (GENCODE v31) with false discovery rate-adjusted P- value < 0.05, the top 5 genes with the lowest P-values are provided in the table. All 4 pts in the “good” group were classified into BM2 and pts in the “poor” group were classified into BM1 (8/22) and BM2 (14/22) (p = 0.07). In addition, all but 2 pts were classified into CMS4. These two pts belong to the “poor” group with one of them classified into CMS1 and the other into CMS2.
Conclusions:
These gene expression analyses suggest that BM2 subtype could be a predictive marker for the efficacy of eribulin and some genes could be novel targets with the goal to improve prognosis of pts with BRAF V600E mt mCRC. This is the first finding for a potential biomarker in this subgroup using RNA-seq analysis tools. These findings will require additional validation. Clinical trial information: UMIN000031552.